Large-scale three-dimensional genomic mapping of heart failure in a European population
We published the first high-precision three-dimensional genomic map of human DCM and normal heart (H3K27ac HiChIP) and combined Hi-C, ChIP-seq, ATAC-seq, RNA-seq and other multi-omics data, which revealed the role of chromatin three-dimensional structural remodeling in the occurrence of DCM, and provided a new target for the future control of DCM and heart failure and the The study is of the following significance.The important findings of this study are as follows: i) A large number of aberrant enhancer-promoter interactions exist in DCM and are significantly associated with the upregulation of early cardiac developmental gene transcription; a class of non-transcribing promoters never reported in the past can act as enhancers to regulate the transcription of DCM-specific genes. ii) The enhancer/promoter regions of DCM-specific interactions have ATAC-seq regions, which can be used for the transcription of DCM genes. iii) The ATAC-seq region of DCM is a region of DCM that can be used for the regulation of DCM. (ii) The ATAC-seq signals of the DCM-specific enhancer/promoter regions were not significantly changed, which further suggested that DCM-specific gene transcription was not achieved by regulating chromatin openness, but by regulating remote chromatin interactions. iii) HAND1 was significantly enriched in the enhancer/promoter regions of the DCM-specific interactions, and overexpression of HAND1 induced enhancer-promoter interactions of cardiac developmental genes and the formation of DCM phenotype. iv) HAND1 was significantly enriched in the DCM-specific enhancer/promoter regions.
 Three-dimensional genome remodelling activates the transcription of early cardiac developmental genes leading to heart failure
Three-dimensional genome remodelling activates the transcription of early cardiac developmental genes leading to heart failure
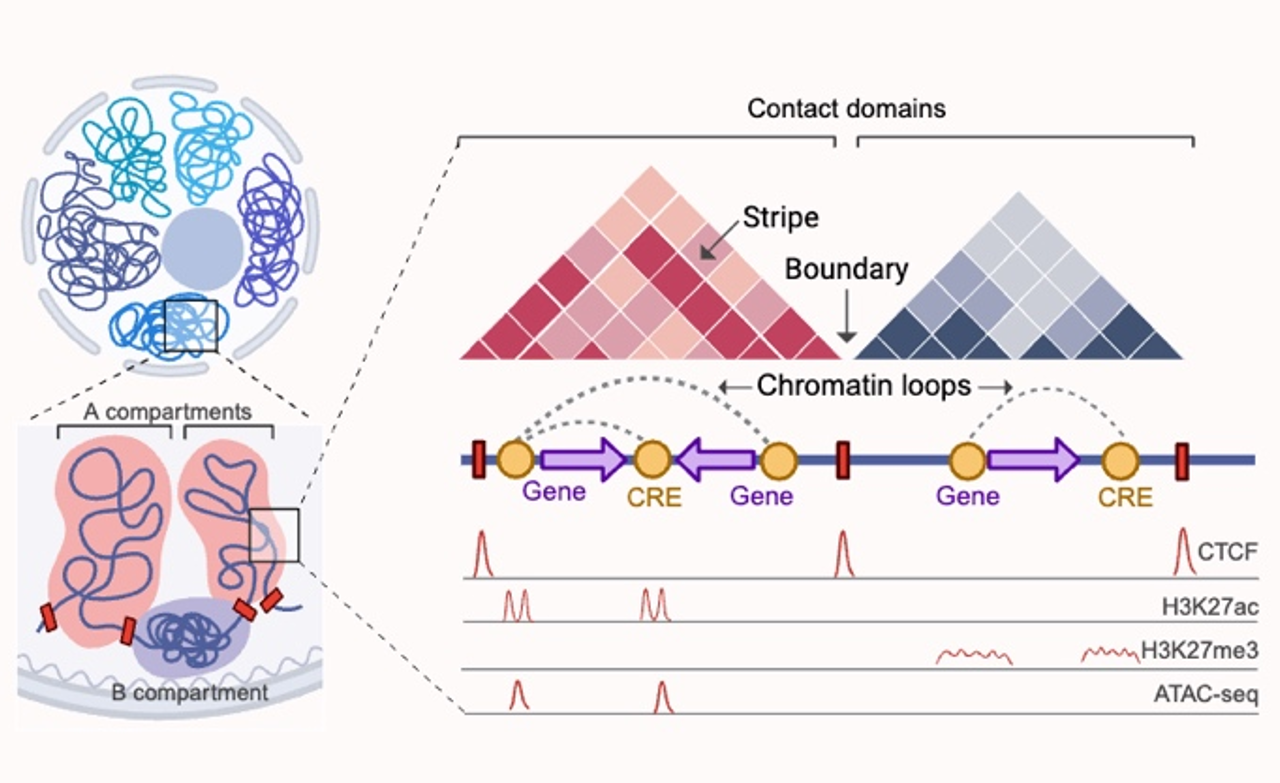
A. Schematic diagram of the experimental design of the DCM three-dimensional genome mapping project. B. Demonstration of the DCM-specific three-dimensional genome structure. C. Schematic diagram of the mechanism by which three-dimensional genome remodelling regulates the occurrence of DCM.
Establishment of a new method for In-situ ChIA-PET 3D genome sequencing
Under the guidance of Prof. Yijun Ruan (a pioneer in the field of 3D genome, former Director of Genome Science Department of Jackson Laboratory, and now Principal Investigator of Institute of Life Sciences, Zhejiang University), we have established a new generation of chromatin 3D genome structure capture technology, In-situ ChIA-PET, together with Dr. Ping Wang (now Research Associate Professor of Northwestern University), etc. Compared with the previous ChIA-PET method established by Prof. Ruan, the amount of cells required has been reduced by nearly 40 times (from 2x10 8 to 5x10 6), and the data quality has been greatly improved, which is of great significance for the analysis of 3D genome structure of primary cells.
We also participated in the establishment of ChIA-PIPE, a data analysis process forin situ ChIA-PET . Subsequently, using small molecule library screening and in situ ChIA-PET , we discovered that bromodomain protein 9 (BRD9) plays an important role in maintaining cell stemness by regulating enhancer-promoter interactions . In addition, we have developed a new 3D genome visualization and analysis tool, EXPRESSO, which integrates the most comprehensive 3D genomics and epigenetics data available for download and viewing to date.
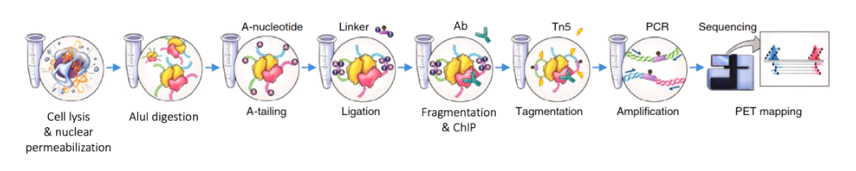 In-situ ChIA-PET Experimental Flowchart
In-situ ChIA-PET Experimental Flowchart
 BRD9-SMAD2/3 Orchestrates Stemness and Tumorigenesis in Pancreatic Ductal Adenocarcinoma
BRD9-SMAD2/3 Orchestrates Stemness and Tumorigenesis in Pancreatic Ductal Adenocarcinoma
 Feng Lab
Feng Lab